Alessandra Bonanni
Senior Lecturer, Division of Chemistry and Biological Chemistry

| Education: |
M.Sc., “Sapienza” University of Rome (Italy) Ph.D., Autonomous University of Barcelona (Spain) |
| Phone: | (+65) 6316 8757 |
| Email: | a.bonanni@ntu.edu.sg |
| Office: | SPMS-CBC-04-19 |
Research Interests
Biosensor technologies
Biosensors are analytical tools which combine a biorecognition agent which provides selectivity and a transducer that confers sensitivity and can convert the biorecognition event into a measurable electronic signal. The advantage of using biosensor over traditional techniques is represented by their low costs, small dimensions, portability, and fast response. The different areas for potential biosensor applications are mainly medical diagnosis, environmental monitoring and food analysis.
DNA analysis
DNA analysis is of extreme importance to solve problems of different nature such as investigation on genetic diseases, detection of food and water contamination by microorganism, studies on breeding origins or tissue matching, and solution of forensic issues. The techniques for genome identification are nowadays mainly based on DNA sequencing by using fluorescent labels and optical detectors. They require a few days for the analysis to be completed and the costs are prohibitive to the general public. Demand has never been greater for innovative technologies which can provide fast, inexpensive and reliable genome information.
The focus of my research is on the development of analytical tools (biosensors) for the rapid, reliable and low cost identification of DNA sequences. In order to achieve that, different electrochemical techniques are used for the detection of the analytical signal. The developed biosensors are based on various platforms (such as gold, carbon and silicon) and different nanomaterials (i.e. gold nanoparticles, graphene nanoplatelets and several quantum dots) are employed for the amplification of the obtained signal and the improvement of detection limit.
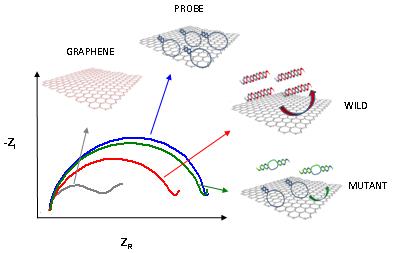
The final step of this work is the application of the developed genosensor to real sample analysis. Once this second part is successfully accomplished, the analytical tool could be eventually integrated into a DNA amplification process, resulting in a portable device for point-of-care diagnostic tests and for very sensitive detection of SNPs correlated to different diseases.
Food Analysis
This field includes research in the two following areas:
- Application of DNA genosensors to identify of DNA sequences correlated to bacteria involved in food contamination (i.e. Salmonella spp, Listeria monocytogenes, Escherichia coli)
- Development of electrochemical biosensors for the detection of antioxidant capacity of food and beverages
Selected Publications
- A. Bonanni, C. K. Chua, G. Zhao, Z Sofer, M. Pumera Inherently Electroactive Graphene Oxide Nanoplatelets as Labels for Single Nucleotide Polymorphism Detection ACS Nano, 6 (2012) 8546-8551
- A. Bonanni, A. Ambrosi, M.Pumera On Oxygen Containing Groups in Chemically Modified Graphenes Chem-Eur. J., 18 (2012) 4541-4548
- A. Bonanni, A. Ambrosi, M.Pumera Nucleic acid functionalized graphene for biosensing Chem-Eur. J., 18 (2012) 1668-1673
- A. Bonanni, M. Pumera Graphene Platform for Hairpin-DNA-Based Impedimetric Genosensing ACS Nano, 5 (2011) 2356-2361
- A. Bonanni, M. Pumera, Y. Miyahara Rapid, Sensitive and Label-free Impedimetric Detection of Single Nucleotide Polymorphism Correlated to Kidney Disease Anal. Chem., 82 (2010) 3772-3779